Insights into the Enhancement of the Poly(ethylene terephthalate) Degradation by FAST-PETase from Computational Modeling
Abstract
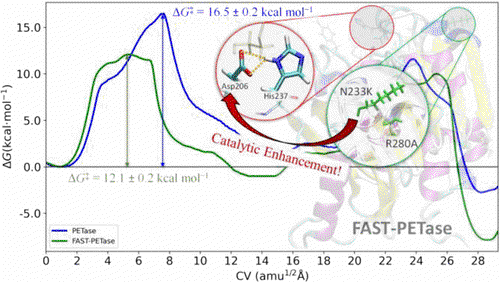
In this work, we investigated the enzymatic biodegradation of polyethylene terephthalate (PET), the most abundant polyester plastic widely used in textiles and packaging but also one of the most discarded single-use plastics. Recent interest in PET biodegradation has grown due to the discovery and engineering of PETase-like enzymes, such as FAST-PETase, which exhibit enhanced depolymerization efficiency. However, the molecular origin of FAST-PETase’s superior activity remains unclear. To address this, we conducted a comprehensive computational study using classical and hybrid quantum mechanics/molecular mechanics (QM/MM) molecular dynamics (MD) simulations. Our results reveal that the rate-limiting step for FAST-PETase is the acylation stage, with an estimated free energy barrier of 12.1 kcal mol⁻¹, significantly lower than that of wild-type PETase (16.5 kcal mol⁻¹). This lower barrier explains FAST-PETase’s enhanced catalytic activity. We attribute this improvement primarily to the N233K mutation, which, despite being located relatively far from the active site, induces a chain folding that affects the positioning of Asp206 in the catalytic triad. This conformational change prevents Asp206 from forming effective hydrogen bonds with neighboring residues, increasing its basicity and enhancing its interaction with the protonated His237 in the transition state of acylation. These effects collectively reduce the catalytic barrier and accelerate PET degradation.