Research projects
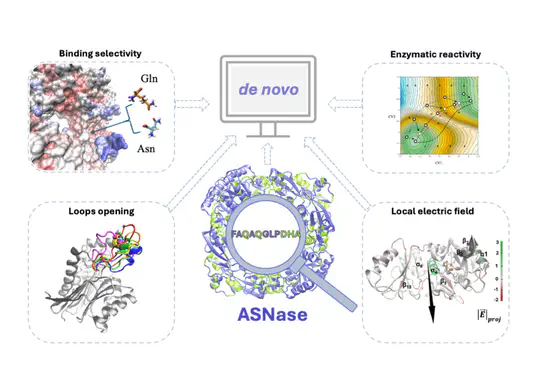
This project focuses on the study of different types and origins of L-asparaginase to enhance their therapeutic potential in leukemia treatment.
The project aims to unravel the chemical mechanism of N-glycosylation in oligosaccharyltransferase (OST) complex, a crucial post-translational modification that influences protein folding, stability, and function.
This study focuses on the enzyme Orotidine 5’-Monophosphate Decarboxylase (OMPDC), which catalyzes the decarboxylation of OMP to UMP in the pyrimidine biosynthesis pathway.
In our research group, we develop machine learning potentials (MLPs) to perform molecular dynamics simulations with ab initio accuracy.
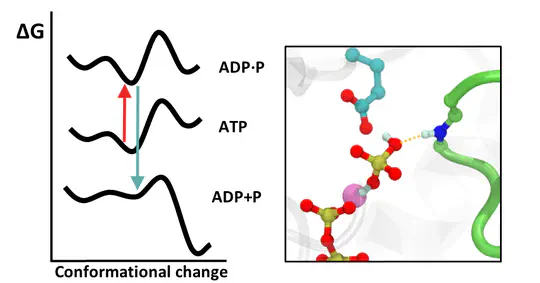
We investigate how the hydrolysis of nucleotides, such as ATP, drives conformational changes in biomolecular motors, particularly in the helicase enzyme.
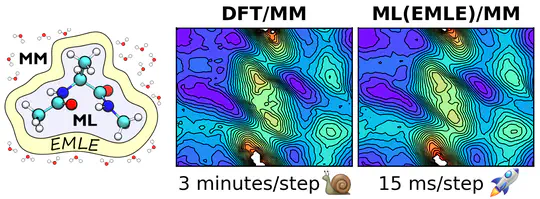
The project focuses on integrating machine‐learned potentials into hybrid QM/MM simulations via electrostatic machine learning embedding (EMLE) approach.
In the 2020 PRACE Fast Track Call for Proposals our group was awared with computational resources to help mitigate the impact of COVID-19.
This project employs computational methods to investigate the intricate mechanism of GTP hydrolysis in G-proteins and the conformational changes that drive their regulatory function.
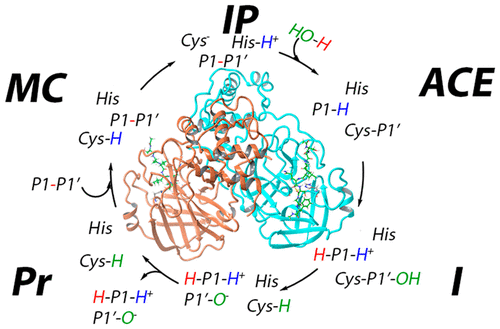
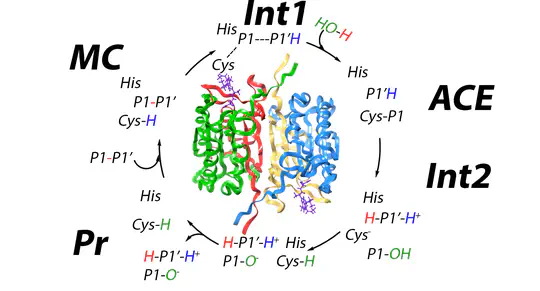
Caspase-1 plays a crucial role in inflammation and immune response, making it a key target in drug development for conditions such as autoimmune diseases and neuroinflammation.