ML/MM embedding with EMLE
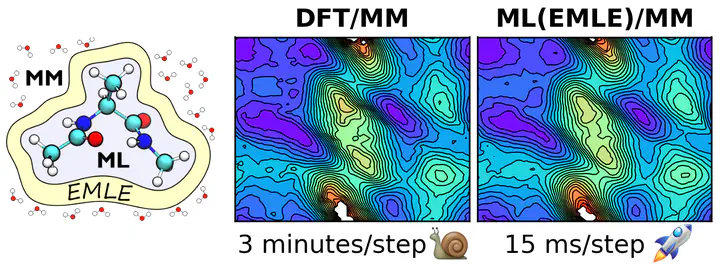
The project focuses on integrating machine‐learned potentials into hybrid QM/MM simulations via electrostatic machine learning embedding (EMLE) approach.
By decoupling the in‐vacuo energy prediction from the environmental polarization effects using physics‐based models for electronic density, charge equilibration, and atomic polarizabilities, the project enables accurate ML potentials to be employed in complex molecular simulations. This framework not only improves the accuracy of energy and force predictions for ground- and excited-state processes but also facilitates seamless incorporation into existing QM/MM software, as demonstrated in applications ranging from small-molecule systems to biomolecular environments. Furthermore, the project led to the development of the emle-engine package, a flexible and open-source tool that implements the ML/MM electrostatic embedding scheme to enable efficient multiscale molecular dynamics simulations.